DS7003 Advanced Decision Making: Predictive Analytics & Machine Learning Assignment Sample
Essential information
Welcome to this M-Level module on Advanced Decision Making. The module forms part of the MSc in Data Science and the Professional Doctorate in Data Science. This guide provides details of scheduled meetings, aims and learning outcomes, approaches to learning and teaching, assessment requirements, and recommended reading for this module. You will need to refer to it throughout the module. Further material may be distributed during the course of the module via Moodle (https://moodle.uel.ac.uk/).
You should consult the relevant Programme Handbook for details of the regulations governing your program.
The Data Science curriculum, for practical reasons, segments the learning process and preparing for research into a number of modules, and those modules into sessions. These sessions will necessarily overlap as their content can be treated from different perspectives and positions. Their content should be treated holistically rather than piecemeal.
Aims and learning outcomes
This module aims to develop a deep understanding of ways of making decisions that are based strongly on data and information. Particular focus will be on mathematical decision-making models including some use of computer-based support. Various case studies will be examined.
Learning outcomes for the module are for students for assignment help
1. Understand the design of decision-making models.
2. Understand the mathematical logic basis of decision-making.
3. Understand how to assign probabilities to uncertain events; assign utilities to possible consequences and make decisions that maximize expected utility.
4. Use of software-based decision-making tools.
5. Critically evaluate alternative decision models.
6. Conduct decision-making exercises.
7. Critically evaluate and analyze data.
8. Compose decision-making based reports.
The definitive description of the module is given in Annex 1.
Approaches to learning and teaching
This module is structured around 10 on-campus sessions over the one-week block.
This module employs a variety of approaches to learning and teaching, which include staff-led presentations, class and small-group discussions, debates, reflection, group exercises, journal-keeping and independent study. You are expected to spend time outside classes preparing for sessions by reading, thinking and engaging in other on- line activities as advised.
Important transferable skills for students intending to undertake or make a career in research include time management, communication skills and evaluation skills. This module places emphasis on developing these skills within the framework of completing the assignment. Strong emphasis is placed on your learning from experience. You must bring a laptop to each class for the practical exercises.
Reading
You are expected to read widely as part of your work for this module, and to spend approximately 15 hours per week in independent study and preparation for assessment. An indicative reading list is given in the Unit Specification Form in Annex 1 of this Guide and additional resources are made available through Moodle. These resources do have considerable overlap between the topics and in some cases repeat some of the knowledge. That is inevitable when looking at a rich literature. You are expected to identify further resources relevant to your particular interests and to the assignment through the Library’s on-line databases and from current affairs materials as Decision Making is very topical.
Attendance
Attendance at sessions is monitored. You should use your student ID card to touch in at the beginning of each session. You do not need to touch out.
Assessment
The assessment for this Module consists of one item: an analysis and review of decision-making using data set(s) of the student’s choosing using machine learning in R for classification, regression or time series of 5,000 words equivalence.
The piece of coursework should be submitted on-line using Moodle. Your work should first be submitted to Turnitin to guard against plagiarism and you can re- submit corrected work up to the due date and time that is specified for each piece of coursework. It is the student’s responsibility to ensure that coursework has been properly submitted on-line to the designated Moodle drop box. Late work may not have accepted for marking and could be treated as a non-submission. If there are justified extenuating circumstances, then these should be submitted on the requisite form according to the general academic regulations. The minimum pass mark for the module is 50%. Students who are unsuccessful in any M-level assessment can be re-assessed once without attendance and should they not be successful on the second take must re-take the module with attendance.
Solution
Introduction
Datasets these days offer several hidden useful information. Data mining is the field of extracting useful data with the help of different algorithms and techniques that covers classification, association, regression and association. It is noteworthy that health care industries have been majorly dependent on data mining tools and techniques. The patients’ risk factors can be predicted if a genuine dataset is examined over these data mining methods. Other than heart diseases, data mining is extensively used to predict other health conditions such as asthma, diabetes, breast cancer, etc. Many well-known studies have been published that proves the efficiency of algorithms such as support vector machine, logistic regression, naïve Bayesian classifier, neural networks and decision trees. The health care research in direction to predict underlying disease is widely in practice since few decades now (Jabbar, 2016).
Cardiovascular diseases are now becoming more chronic in last few years. Every year millions of people including all age groups die of severe heart diseases. The disease might not be detected at early stage due to which people have been dying as soon as the disease is diagnosed. However, early detection of heart disease can save patients’ lives as their risk factors are known before the time has passed. This prediction at early stage may help in minimizing mortality rates. The heart diseases are of various kinds that includes coronary heart disease, cardio myopathy, hypertensive heard disease, heart failure, etc. These conditions are caused due to diabetic conditions, hypertensions, high cholesterol levels in body, obesity, etc.
Data mining methods and techniques can be fruitful in early prognosis of disease that can calculate high risk factors by analyzing existing dataset. The data mining tools and algorithms are proven to be accurate enough to predict right information (Gandhi, 2015). The dataset used in this research on which analysis is done is collected from Cleveland Heart Clinic; the database is popularly known as Cleveland Heart Disease Database (CHDD) used by many researchers. The information about its key attributes is discussed later in the chapter.
The research report is organized as follows. The next section offers machine learning insights and details of categories of algorithms prevalent in machine learning. Later, literature is presented towards the researches being conducted to predict cardiovascular diseases by other researchers. Then, methodology is discussed about the work conducted in this research. It provides detailed description of how diseases are predicted by support vector machine and logistic regression. Additionally, both the techniques are tested for their accuracy and hence compared. The objectives of this research are two-fold: First, finding how accurate these two selected methods are for predicting heart disease. Second, comparison of two methods based on their accuracy.
Traditionally, statistical techniques were used to develop intelligent applications. The predictions were simply based on interpolation formulae as learned in high schools. However, in today’s AI applications, traditional interpolation formulae could fail; hence, new techniques are successfully developed to fulfill current AI needs and requirements. Most popular methods in artificial intelligence and machine learning are regression, clustering, classification, probability and decision trees. These methods contain several different algorithms to suit different class of problems to solve complex problems (Babu, 2017). Machine learning is further categorized as in figure 1.
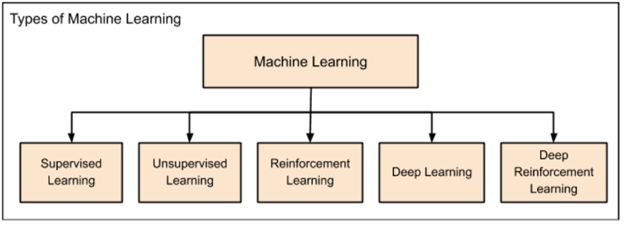
Figure 1 Categories of Machine Learning (Babu, 2017)
The categories of machine learning are:
Supervised Learning: The very first found machine learning technique used first to predict residential pricing. The supervised learning, as the name suggests, supervises entire application working. Just as child is taught to walk, by holding hands, the supervised learning algorithm monitors each and every step of application to achieve best results. Regression and Classification are kinds of methods used in supervised learning.
Regression: The machine is trained first by providing results for the input value. The machine then learns the relationship between input and output values. Once the machine learns it, the value of output is predicted for input provided based on empirical relationship between previous inputs and their respected outputs.
Classification: The classification problem efficiently categorizes the pool of data into predefined classes. For example, if a box contains blue, red and green balls; the classification algorithm can sort and find the number of balls of each color in the box. The classification algorithm classifies objects of same kind into one category whose label is previously named. This technique of machine learning falls under supervised learning where group names are already defined. Objects are classified as per nature described.
Unsupervised Learning: This method doesn’t require any other agent to supervise it. Completely opposite to supervised learning, unsupervised learning technique never allows machine to train itself on the basis of known outputs from certain inputs. Rather, the algorithm forces machine to find all possible values of the given input. This is achieved by identifying the features mostly appearing in input.
Reinforcement Learning: The idea is to reward the machine whenever it achieves desired results. Consider at an instance, when training a dog to fetch a ball. If a ball thrown is fetched by dog, it is rewarded by the master. This helps dog to understand how to do the commanded task. This type of learning is termed as reinforcement. This algorithm is usually applied in games. The application should analyze the possible next moves after each next step. If the predicted next step is correct, the machine is rewarded else it is punished or penalized. This will help algorithm to identify right and wrong moves; hence the machine will learn right moves as the game proceeds. Thus, the accuracy towards winning keeps on increasing.
Deep Learning: To aid in analyzing big data, deep learning was introduced so that applications can be designed as human brains that can create artificial neural network.
Deep Reinforcement Learning: Similar to reinforcement learning, when deep learning application or machine works the same as expected, it is rewarded in deep reinforcement learning.
These algorithms are founded as listed from supervised learning to deep reinforcement learning. With need to solve each new complex problem, new machine learning method was introduced.
Literature Review
Researchers have been trying since decades the accurate way to predict heart diseases so that lives of millions of people can be saved. Authors have developed a system to detect heart diseases accurately by monitoring heart rate data collected from a survey (Wijaya, 2013). The tools act as a data feeder and retriever in the survey which is a pitfall. In other words, if tool isn’t available, data cannot be read. However, the correct prediction of severity of heart disease could increase person’s life span by developing healthy lifestyle. The system developed by authors (Gavhane, 2018) is less accurate as results obtained. The other researchers (Ul Haq et al., 2018) developed a system which was comparatively reliable that included usage of classification technique. The back propagation algorithm was used to predict heart diseases; and the results were promising (Xing et al., 2007). Used feature selection for gaining more accurate results. The authors developed a system to find heart diseases, cancer and diabetes from the dataset.
Apart from usage of feature selection and back propagation few researchers have developed systems that made good use of other algorithms such as support vector machine, neural network, regression, decision trees and several classifiers such as naïve bayes. The datasets were different and common for few authors but the techniques were unique. The datasets used were collected all around the globe; the results were different and accuracies differed in many ways as use of techniques were different. Xing et al. collected dataset of thousand patients wherein different parameters were included such as hypertension, smoking habits, age, gender, diabetic or not, etc. The authors developed three algorithms: SVM, ANN and decision trees. The accuracies calculated were 92.1%, 91.0% and 89.6% respectively.
Chen et al. developed a system to predict heart diseases with the help of algorithms like SVM, neural network, Bayes Calssifier, decision trees and logistic regression. The most accurate of all is support vector machine (90.5%) whereas the accuracy of logistic regression was the least (73.9%). Neural network, Bayes classifier and decision trees had accuracies 88.9%, 82.2%, 77.9% respectively.
Many researchers have selected different attributes of dataset for several machine learning techniques. It was observed that it does not provide a justified basis to compare the techniques for their accuracy. With this thing in consideration, Soni et al. selected those attributes that can affect largely to heart of patients that consist of gender, obesity, hypertension, smoking habits, drinking habits, salt intake, cholesterol level, diabetes, family history of the same disease, no physical exercises, etc. Similarly, Shouman et al. identified blood pressure as another risk factor that can be considered while predicting heart disease. Cleveland Heart Disease database is highly considered by researchers as it is one of the most trusted dataset for analysis. The very first research on this dataset was conducted in 1989 by Detrano et al. where logistic regression was implemented and accuracy of 77% was attained. This is worth noting that accuracy of models after their evolution is dramatically increasing and it is found to be above 90%. Many researchers have used combination of machine learning techniques to attain accuracy as high as possible.
Other than SVM and logistic regression, k-means clustering and c4.5 decision trees were implemented by Manikanthan and Latha to predict heart disease. The accuracy of the system was 92% which was considered as a promising approach. Nor Heiman et al. studied methods in decision tree algorithm to minimize errors. Dota et al. used decision tree algorithm to compare accuracies to detect quality of water. Rajeshwari et al chose some attributes that can help patients to find the list of medical tests that are of no need. Lakshmi et al used many data mining techniques such as c4.5, k-means, decision trees etc. and found their specificity. It was observed that PLS-DA shown promising results in prediction of heart disease. Taneja used Naïve Bayes classifier to choose attributes from dataset. He proved that data mining techniques for feature selection are equally important as that of prediction or classification itself. Sudhakar and Manimekalai comparatively presented many data mining techniques as a survey research paper. Ahmed and Hamman performed data mining for cardiovascular predictions using support vector machine and clustering techniques. They also performed association rules mining for attribute selection. Sugumaran et al performed data mining for fault detection using svm and classification.
Methodology
The research concentrates on implementation of logistic regression and support vector machines on Cleveland heart disease dataset in order to find and compare their accuracies.
Support Vector Machine
A machine learning model that helps in finding known patterns for classification and regression. SVM is mostly applied when the data can be classified into two groups. The hyper plane generated by SVM can be useful in partitioning similar kind of objects with other quite efficiently. It is considered that the distance between two classes should be maximum to prove that the model is good. To solve real world problems SVM is highly considered since it is a pure mathematical model and may provide best results. It is also better to SVM when a dataset consists of many attributes. The foremost task, an SVM does is mapping data into kernel space. Selection of kernel space can be quite challenging with SVM.
Logistic Regression
Logistic regression can predict the outcome with limited group categorical value dependent on a variable. In CHDD, the outcome should be heart disease detected or not. The outcomes are usually binary in logistic regressions. With binary, it means having two values at maximum. The probability of success is largely measured by logistic regression.
The system flowchart is presented below in figure 2. The Cleveland heart disease dataset is used via its link. The next step is data exploration and pre-processing. To accomplish this, features are selected first, in this dataset important parameters are first identified and selected for further processing. Support Vector Machine model is first applied and its accuracy is stored in an array. Later, Logistic Regression model is also applied and its accuracy is again stored in the same array. Finally, the accuracy of both the models applied is compared and results are presented. The experimental analysis is presented in next section.
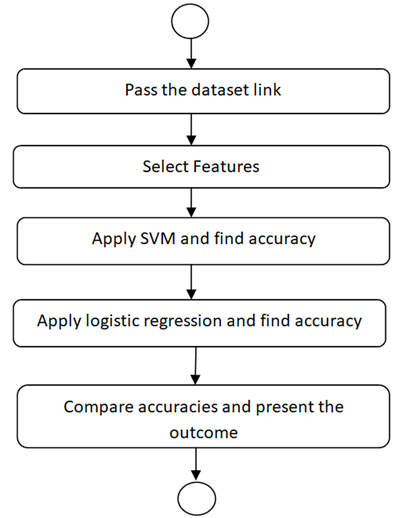
Figure 2 Flowchart of Proposed Research
Data Exploration using R
Data Exploration is the initial and the most important step of data mining process. It not only helps in finding uncovered aspects but also offers details of the data under observation. It helps in building a system that can be accurate to its peak.
Data Exploration is a stepping stone for analysts to visualize dataset by applying statistical methods on dataset. The data and its nature are well understood by data exploration step. The size, type of attributes, format of attributes plays important roles in data exploration. The manual and automated, both ways are widely used as per data analysts ease. This step helps them to visualize important graphs and hence find correlation between attributes. The outlier detection and distribution of values of attributes can be performed by data exploration tools much efficiently than that of manual efforts. The datasets collected by other researchers are raw data and to find few important insights, it is necessary to perform data exploration. The data collection is a pillar to data exploration. To select correct data for analysis is again an important task. While selecting a dataset analyst must know what are the important attribute types to lookup.
Data exploration need is quite well justified. It is always observed that data in the form of pictures, graphs and any form of visualization is more easily grasped by viewers than that of numerical data in tabular form or in excel sheets. Hence, it is a tedious task for analysts to put millions of data rows and their corresponding attributes to frame in a graph so that it is well understood to the viewers.
Data visualization in R and its exploration is quite simple as it takes syntax to learn. It first starts with loading dataset chosen into allowable extensions such as csv, xlx, json and txt. The variables are converted into characters from numbers to aid the parsing in R. A dataset is then transposed from wide to narrow structure. Dataset is sorted accordingly and plots are created as per visualization requirements. Duplicate variables can be easily removed using R. Merging and joining dataset as well as count and sum functions are also easily available. Data Exploration in R is based on fixed syntax as presented next.
1. Reading a dataset

2. Transpose a dataset

3. Remove duplicate values

4. Create a histogram
The operations provided here are just few from the pool of operations in R studio. There are numerous libraries one can import and use different function already defined in it.
Data Source
Dataset: Heart Disease Dataset from UCI Repository
Link: https://archive.ics.uci.edu/ml/datasets/Heart+Disease
The heart disease dataset is a collection of 303 records identified by 76 attributes. Out of which, goal attribute identifies if there is presence of heart disease in the underlying patient. It is represented by an integer value, where 0 indicates no heart disease and presence of heart disease is classified as values 1 to 4; low to the scales of severe.
14. #58 (num) (the predicted attribute)
Complete attribute documentation:
1 id: patient identification number
2 ccf: social security number (I replaced this with a dummy value of 0)
3 age: age in years
4 sex: sex (1 = male; 0 = female)
5 painloc: chest pain location (1 = substernal; 0 = otherwise)
6 painexer (1 = provoked by exertion; 0 = otherwise)
7 relrest (1 = relieved after rest; 0 = otherwise)
8 pncaden (sum of 5, 6, and 7)
9 cp: chest pain type
-- Value 1: typical angina
-- Value 2: atypical angina
-- Value 3: non-anginal pain
-- Value 4: asymptomatic
10 trestbps: resting blood pressure (in mm Hg on admission to the hospital)
11 htn
12 chol: serum cholestoral in mg/dl
13 smoke: I believe this is 1 = yes; 0 = no (is or is not a smoker)
14 cigs (cigarettes per day)
15 years (number of years as a smoker)
16 fbs: (fasting blood sugar > 120 mg/dl) (1 = true; 0 = false)
17 dm (1 = history of diabetes; 0 = no such history)
18 famhist: family history of coronary artery disease (1 = yes; 0 = no)
19 restecg: resting electrocardiographic results
-- Value 0: normal
-- Value 1: having ST-T wave abnormality (T wave inversions and/or ST elevation or depression of > 0.05 mV)
-- Value 2: showing probable or definite left ventricular hypertrophy by Estes' criteria
20 ekgmo (month of exercise ECG reading)
21 ekgday(day of exercise ECG reading)
22 ekgyr (year of exercise ECG reading)
23 dig (digitalis used furing exercise ECG: 1 = yes; 0 = no)
24 prop (Beta blocker used during exercise ECG: 1 = yes; 0 = no)
25 nitr (nitrates used during exercise ECG: 1 = yes; 0 = no)
26 pro (calcium channel blocker used during exercise ECG: 1 = yes; 0 = no)
27 diuretic (diuretic used used during exercise ECG: 1 = yes; 0 = no)
28 proto: exercise protocol
1 = Bruce
2 = Kottus
3 = McHenry
4 = fast Balke
5 = Balke
6 = Noughton
7 = bike 150 kpa min/min (Not sure if "kpa min/min" is what was written!)
8 = bike 125 kpa min/min
9 = bike 100 kpa min/min
10 = bike 75 kpa min/min
11 = bike 50 kpa min/min
12 = arm ergometer
29 thaldur: duration of exercise test in minutes
30 thaltime: time when ST measure depression was noted
31 met: mets achieved
32 thalach: maximum heart rate achieved
33 thalrest: resting heart rate
34 tpeakbps: peak exercise blood pressure (first of 2 parts)
35 tpeakbpd: peak exercise blood pressure (second of 2 parts)
36 dummy
37 trestbpd: resting blood pressure
38 exang: exercise induced angina (1 = yes; 0 = no)
39 xhypo: (1 = yes; 0 = no)
40 oldpeak = ST depression induced by exercise relative to rest
41 slope: the slope of the peak exercise ST segment
-- Value 1: upsloping
-- Value 2: flat
-- Value 3: downsloping
42 rldv5: height at rest
43 rldv5e: height at peak exercise
44 ca: number of major vessels (0-3) colored by flourosopy
45 restckm: irrelevant
46 exerckm: irrelevant
47 restef: rest raidonuclid (sp?) ejection fraction
48 restwm: rest wall (sp?) motion abnormality
0 = none
1 = mild or moderate
2 = moderate or severe
3 = akinesis or dyskmem (sp?)
49 exeref: exercise radinalid (sp?) ejection fraction
50 exerwm: exercise wall (sp?) motion
51 thal: 3 = normal; 6 = fixed defect; 7 = reversable defect
52 thalsev: not used
53 thalpul: not used
54 earlobe: not used
55 cmo: month of cardiac cath (sp?) (perhaps "call")
56 cday: day of cardiac cath (sp?)
57 cyr: year of cardiac cath (sp?)
58 num: diagnosis of heart disease (angiographic disease status)
-- Value 0: < 50% diameter narrowing
-- Value 1: > 50% diameter narrowing
(in any major vessel: attributes 59 through 68 are vessels)
59 lmt
60 ladprox
61 laddist
62 diag
63 cxmain
64 ramus
65 om1
66 om2
67 rcaprox
68 rcadist
69 lvx1: not used
70 lvx2: not used
71 lvx3: not used
72 lvx4: not used
73 lvf: not used
74 cathef: not used
75 junk: not used
76 name: last name of patient
Experimental Analysis
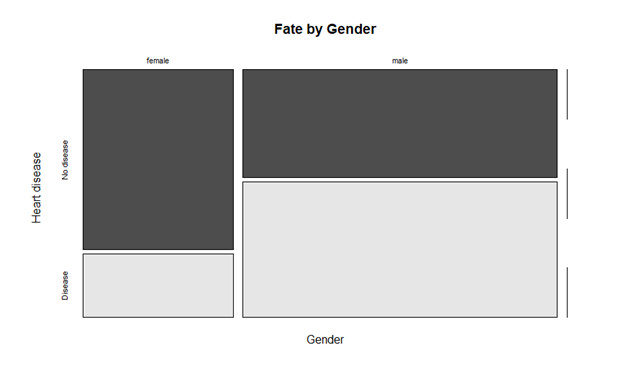
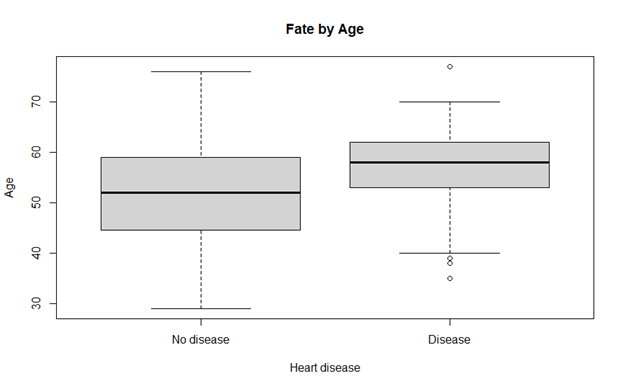
Comparisons
The metrics used to compare SVM and logistic regression is AUC and accuracy. As seen below, support vector machine performs better than logistic regression. Following is the snapshot of accuracy list of both the techniques used.
Some researchers have found that logistic regression performs better in most cases if selection of attribute is slightly different. However, in our prediction, support vector machine performed better. However, there is still need to finding another method that can provide better accuracy than SVM.
Conclusions
The research concentrated on mining data related to heart disease. The dataset is popular and hence highly trustworthy to conduct concrete research on. Only fourteen out of seventy-six attributes were selected to test models named support vector machine and logistic regression. This research explored data in R studio with the help of in built libraries for applying several models with utmost ease. Data mining has now gained huge importance due to curiosity of prediction for necessary information. Today’s world contains gigantic datasets built over the period of years. These datasets are future predictors for many hidden aspects. The models implemented in R shows that accuracy of support vector machine is better than logistic regression. However, the accuracy should be increased by implementing other data mining techniques such as decision trees or naïve bayes classifier, etc. The research report presents details related to machine learning domain along with data exploration in R. R studio is a user friendly platform with fixed syntax to explore data and find useful information from it.
References
Ahmed, A. & Hannan, S. (2012). Data mining techniques to find out heart diseases: An overview. International Journal of Innovative Technology and Exploring Engineering (IJITEE), Volume-1, Issue-4.
Babu, S. (2017). Heart disease diagnosis using data mining technique. International conference on electronics, communication and aerospace technology ICECA2017.
Chen, J. Xi, G. Xing, Y. Chen, J. & Wang, J. (2007). Predicting Syndrome by NEI Specifications: A Comparison of Five Data Mining Algorithms in Coronary Heart Disease. Life System Modeling and Simulation Lecture Notes in Computer Science, pp 129-135.
Detrano, R. Janosi, A. Steinbrunn, W. Pfisterer, M. Schmid, J. Sandhu, S. Guppy, K. Lee, S. & Froelicher, V. (1989). International application of a new probability algorithm for the diagnosis of coronary artery disease. The American Journal of Cardiology, pp 304-310.15.
Dota, M. Cugnasca, C. Savio, D. (2015). Comparative analysis of decision tree algorithms on quality of water contaminated with soil, Ciencia Rural, Santa Maria, v.45, n.2, p.267-273, ISSN 0103-8478.
Gandhi, M. (2015). Prediction in heart disease using techniques of data mining. International conference on futuristic trend in computational analysis and knowledge management (ABLAZE-2015).
Gavhane, A. (2018). Prediction of heart disease using machine learning, ISBN: 978-1-5386-0965-1.
Jabbar, M. (2016). Heart disease prediction system based on hidden Navie Bayes Classifier. International conference on circuits , controls, communications and computing (14C).
Lakshmi, K. Veera Krishna, M. and Prem Kumar, S. (2013). Performance Comparison of Data Mining Techniques for Predicting of Heart Disease Survivability. International Journal of Scientific and Research Publications, 3(6).
Mai Shouman, M. Turner, T. & Stocker, R. (2012). Using Data Mining Techniques In Heart Disease Diagnoses And Treatment. Electronics, Communications and Computers (JECECC), 2012 Japan-Egypt Conference March 2012, pp 173-177.
Manikantan, V. & Latha, S. (2013). Predicting the analysis of heart disease symptoms using medicinal data mining techniques. International Journal on Advanced Computer Theory and Engineering (IJACTE), ISSN (Print) : 2319 – 2526, 2(2).
Nor Haizan, W. Mohamed, M. & Omar, A. (2012). A comparative study of reduced error pruning method in decision tree algorithms. IEEE International conferance on control system, Computing and Engineering, 23 - 25, Penang, Malaysia.
Rajeswari, K. Vaithiyanathan, V & Pede, S. (2013). Feature Selection for Classification in Medical Data Mining. International Journal of emerging trends and technologies in Computer Science, 2(2).
Soni, J. Ansari, U. & Sharma, D. (2011). Predictive Data Mining for Medical Diagnosis: An
Overview of Heart Disease Prediction. International Journal of Computer Applications, doi 10.5120/2237-2860.
Sudhakar, K. and Manimekalai, M. (2014). Study of Heart Disease Prediction using Data Mining, International Journal of Advanced Research in Computer Science and Software Engineering, 4(1), ISSN: 2277 128X.
Sugumaran, V. Muralidharan, V. & Ramachandran, K. (2007). Feature selection using Decision Tree and classification through proximal support vector machine for fault diagnostics of roller bearing. mechanical systems and signal processing, 21 (2), Pages 930-942.
Taneja, A. (2013). Heart Disease Prediction System Using Data Mining Techniques. J. Comp. Sci. & Technol., 6(4), 457-466.
Ul Haq, A. Li, J. Memon, M. Nazir, S., & Sun, R. (2018). A Hybrid Intelligent System Framework for the Prediction of Heart Disease Using Machine Learning Algorithms.
Wijaya, R. (2013). Preliminary design of estimation heart disease by using machine learning ANN within one year. communication technology and electric vehicle technology , bandung-bali, Indonesia.
Xing, Y. Wang, J. & Gao, Z. (2007). Combination data mining methods with new medical data to predicting outcome of Coronary Heart Disease. Convergence Information Technology. International Conference November 2007, pp 868-872.




.png)
~5.png)
.png)
~1.png)






















































.png)






